Our research
Our goal is to better understand how genes regulate each other and how disease disrupts gene networks. This goal is realized by developing software that enables us and others to analyze complex genomic datasets that sample the transcriptional landscape. With this mission in mind, we build models with insight of the experimental protocols and biological questions while utilizing and extending the latest theory of computer science and statistics.
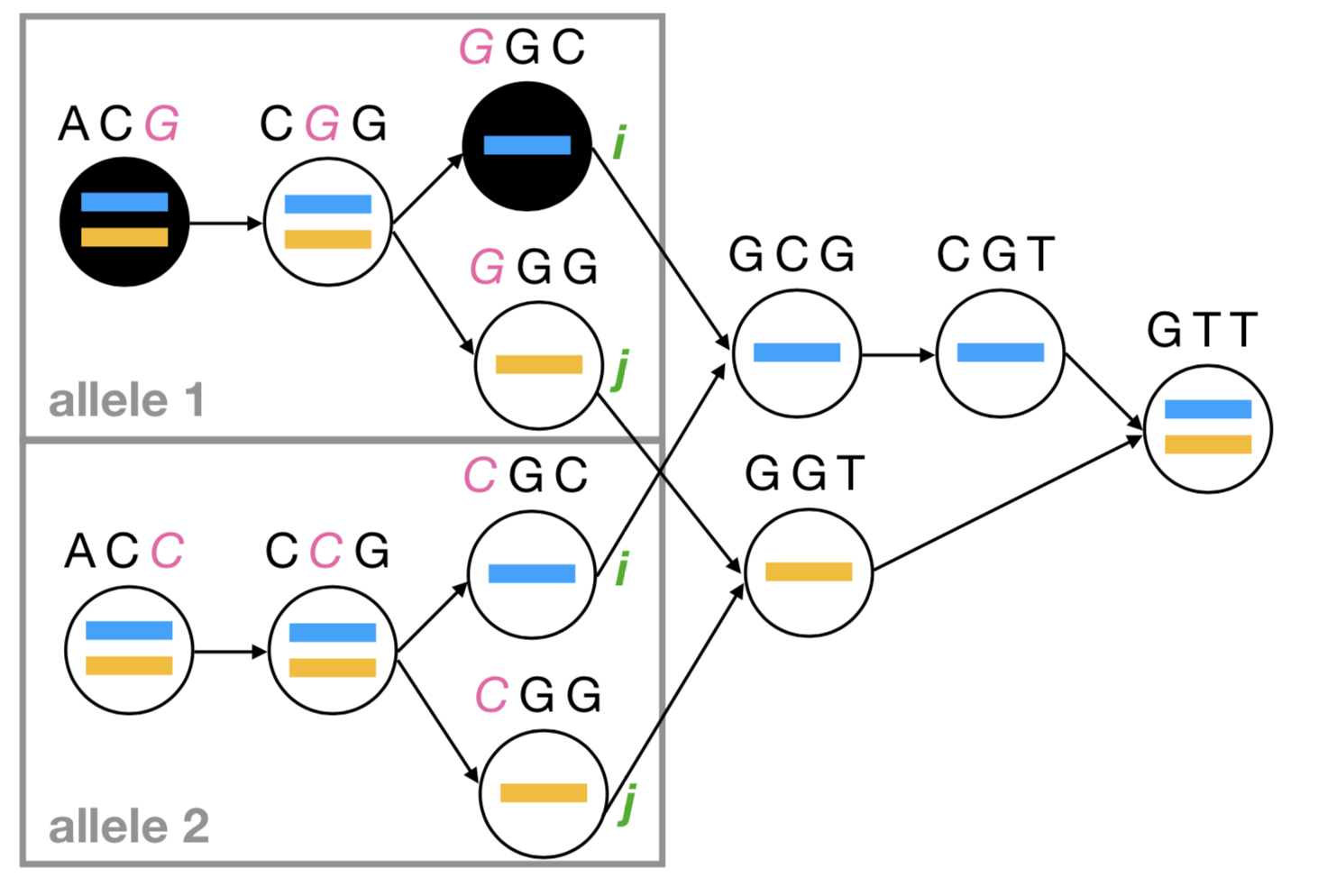
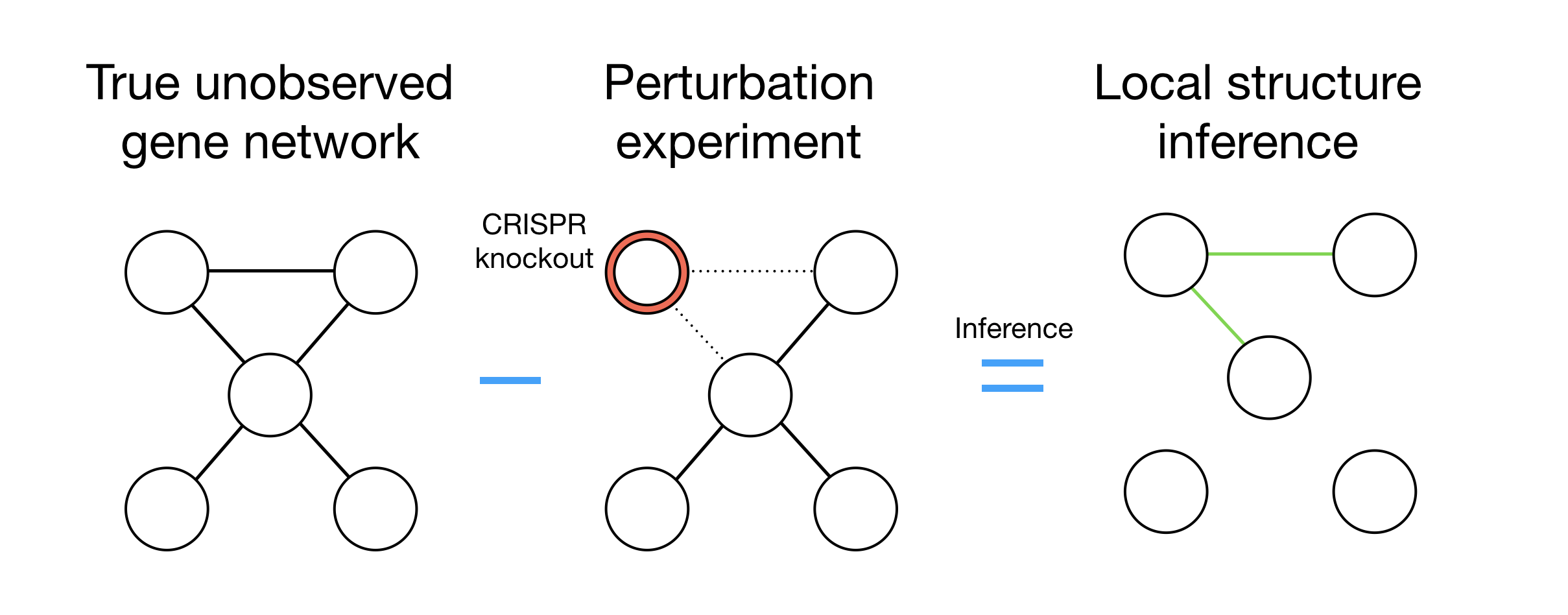
Structure learning for gene regulation
We are utilizing and extending new developments in machine learning to model the behavior of modern genetic perturbations (i.e. CRISPR) to develop tools for inferring gene network structure.
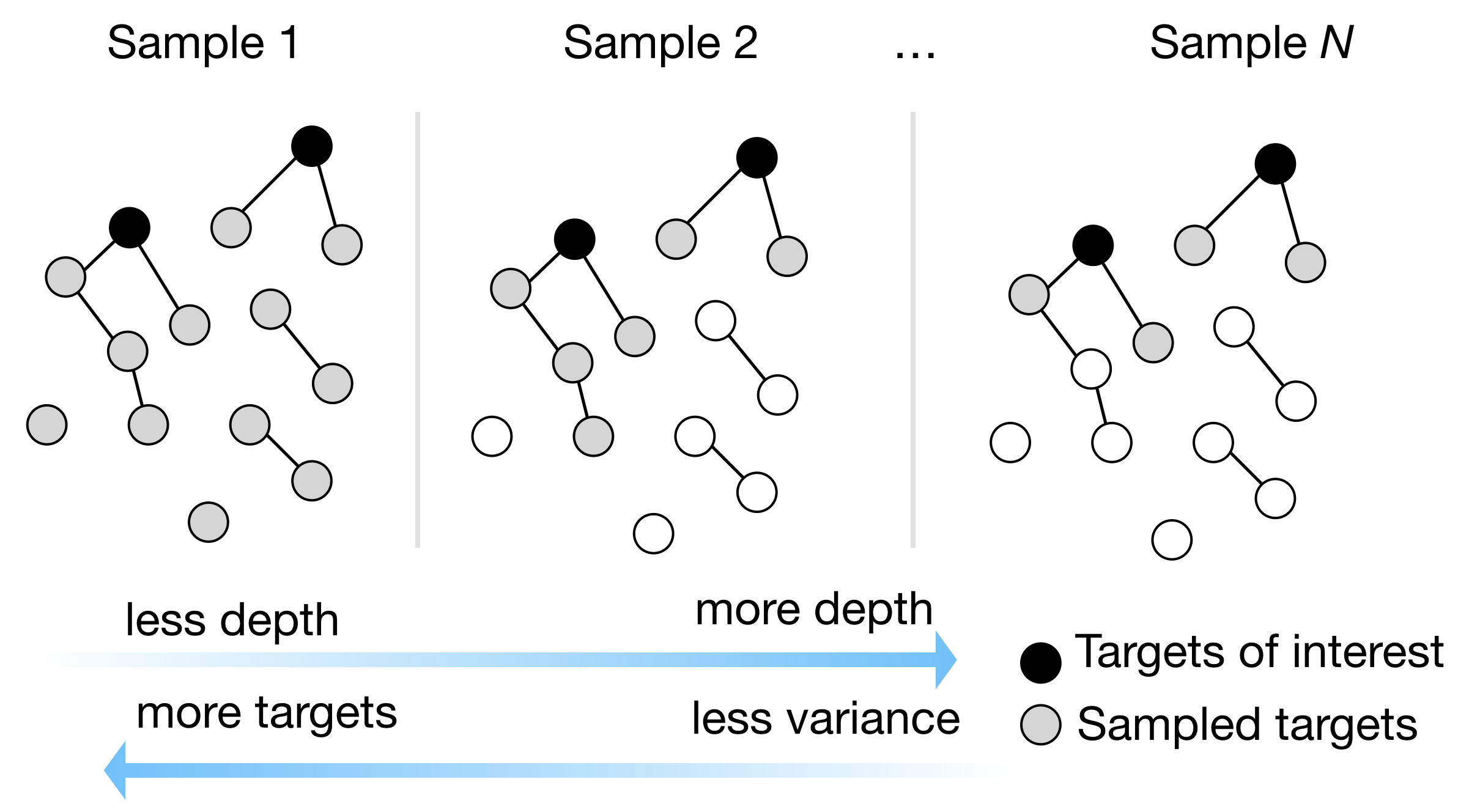
Active learning for experimental design
Modern experiments can capture multiple phenotypes at the cell-level while capturing millions of cells in one library preparation. Seemingly unbounded sampling potential means that naively sampling everything is likely to be prohibitively expensive. We are developing active machine learning techniques to guide experimentalists through iterative sampling towards the most informative data given prior data, goals, and cost.
Quantifying the genetic component of RNA
We are developing methods to integrate the advances in RNA-seq quantification uncertainty, population specific sequence alignment, and causal inference to understand genetic drivers of disease.